Introduction to joint profiling of native and R code
Kirill Müller
2021-03-27
Source:vignettes/jointprof.Rmd
jointprof.RmdThe three rules of software optimization are:
- Don’t.
- Don’t do it yet.
- Profile before optimizing.
A profiler is a tool that identifies which parts of your code take the most time. One way to do this is to run the code and halt execution every so often (by default 50 times per second), and record the call stack on each occurrence. The combined samples will likely show in which part of your code the most time is spent.
In R, a sampling profiler is available via Rprof(), but it provides detailed measurements only for R code. This vignette shows how to include detailed information on native code run times in the profile.
Prerequisite: Code to profile
For this vignette, we’ll be profiling R and C++ implementations of functions that computes the Fibonacci sequence using dynamic programming:
fib_r <- function(x) {
if (x <= 2) return(rep(1, x))
fib <- fib_r(x - 1)
c(fib, fib[x - 1] + fib[x - 2])
}#include <Rcpp.h>
// [[Rcpp::export]]
Rcpp::NumericVector fib_cpp(int x) {
if (x <= 1) return Rcpp::NumericVector::create(1);
if (x <= 2) return Rcpp::NumericVector::create(1, 1);
Rcpp::NumericVector fib = fib_cpp(x - 1);
// Zero-based indexing!
fib.push_back(fib[x - 1 - 1] + fib[x - 2 - 1]);
return fib;
}(A few tweaks are necessary to allow profiling the Rcpp-compiled function when run from the vignette. See the source for details.)
The implementations are supposed to behave identically.
fib_r(10)
#> [1] 1 1 2 3 5 8 13 21 34 55
fib_cpp(10)
#> [1] 1 1 2 3 5 8 13 21 34 55Our test function test() calls both functions and compares their output in a loop:
A minimal example
Rprof() creates a text file that contains the stack trace for each interruption. The summaryRprof() function converts this to a data frame with one row per function and the corresponding run time estimates (self: code in that function only, total: including all descendant functions):
out_file <- tempfile("jointprof", fileext = ".out")
Rprof(out_file, gc.profiling = TRUE)
for (i in 1:10) test(800)
Rprof(NULL)
summary <- summaryRprof(out_file)
summary$by.self
#> self.time self.pct total.time total.pct
#> "fib_r" 4.86 34.66 10.74 76.60
#> "c" 4.74 33.81 5.30 37.80
#> ".Call" 2.76 19.69 3.22 22.97
#> "<GC>" 1.22 8.70 1.22 8.70
#> "-" 0.24 1.71 0.26 1.85
#> "<=" 0.08 0.57 0.08 0.57
#> "+" 0.06 0.43 0.06 0.43
#> "test" 0.02 0.14 14.02 100.00
#> "stopifnot" 0.02 0.14 14.00 99.86
#> "...elt" 0.02 0.14 13.98 99.71This is a nice overview, but for fun_cpp() the run time is hidden in the entry for .Call(): calls into native code are opaque and cannot be resolved further. Enter joint profiling:
library(jointprof)
out_file <- tempfile("jointprof", fileext = ".out")
start_profiler(out_file)
for (i in 1:10) test(700)
stop_profiler()
summary <- summaryRprof(out_file)
summary$by.selfThe summary now also contains a breakdown for native functions below the R code that calls them.
Installation
Ubuntu
Other Linux distributions may work if you install the right system dependencies (let me know which!).
-
Install system dependencies:
-
Install
pprof: -
Install the package:
# install.packages("remotes") remotes::install_github("r-prof/jointprof")
OS X
-
Install system dependencies:
-
Install
gperftools(currently from a branch, pull request pending): -
Install
pprof: -
Install the package:
# install.packages("remotes") remotes::install_github("r-prof/jointprof")
Usage
The start_profiler() and stop_profiler() functions are replacements for Rprof() and Rprof(NULL), respectively. The generated output file is compatible with that generated by Rprof(). This enables a variety of ways to analyze the results, see also the subsequent section.
out_file <- tempfile("jointprof", fileext = ".out")
start_profiler(out_file)
for (i in 1:10) test(700)
stop_profiler()
strtrim(readLines(out_file, n = 12), 90)Furthermore stop_profiler() returns an in-memory representation of the profiled data, invisibly. See ?profiler::validate_profile for a description of the data format.
out_file <- tempfile("jointprof", fileext = ".out")
start_profiler(out_file)
for (i in 1:10) test(700)
profile_data <- stop_profiler()
profile_data
profile_data$samples
profile_data$locations
profile_data$functionsThe profile data can be edited, and written to a file via write_rprof() or (in pprof-compatible format) via profile::write_pprof(). The example below removes the first 15 entries from all samples, and writes the resulting profile data in pprof format:
profile_data$samples$locations <- lapply(
profile_data$samples$locations,
head,
-15
)
pprof_file <- tempfile("jointprof", fileext = ".pb.gz")
profile::write_pprof(profile_data, pprof_file)We then use pprof to visualize the call graph, using find_pprof() to get the path to the pprof executable:
dir.create(knit_dir("jointprof_fig"), recursive = TRUE, showWarnings = FALSE)
svg_file <- knit_dir("jointprof_fig/minimal.svg")
system2(
find_pprof(),
c(
"-svg",
"-nodefraction 0.01",
"-output",
shQuote(svg_file),
shQuote(pprof_file)
)
)
png_file <- knit_dir("jointprof_fig/minimal.png")
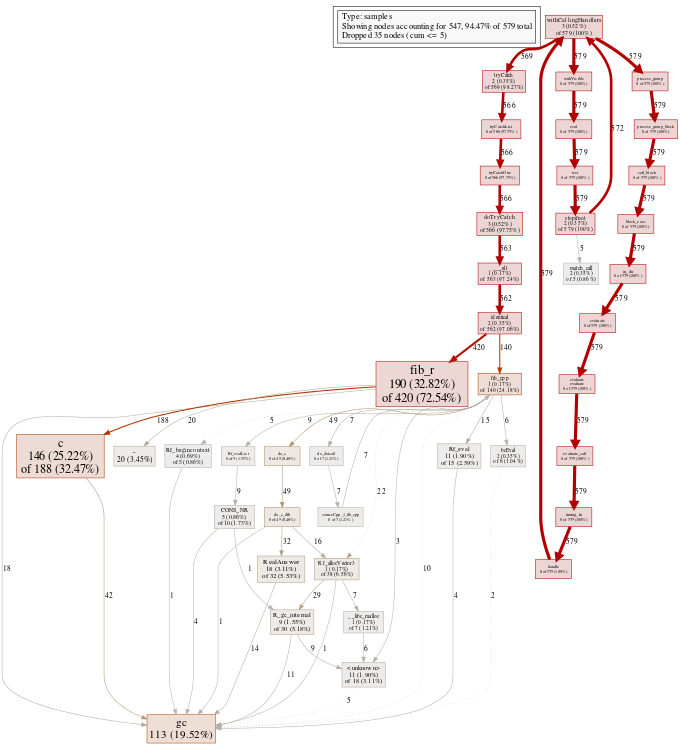
rsvg::rsvg_png(svg_file, png_file, width = 680)The call graph shows a unified view of run time costs for both R and native code. (Click on the image to see the SVG version.)
For interactive exploration of a call graph, try the built-in web browser view:
system2(
find_pprof(),
c(
"-http",
"localhost:8080",
shQuote(pprof_file)
)
)Other packages for analyzing profiler data
I’m aware of the following R packages that process profiler data, ordered by date of first CRAN release:
proftools: Mature, the paper’s includes facilities for summarizing results at the function, call, and source line level; for filtering to narrow the focus to functions of primary interest; and for visualizing profiling data. Also offers an export to callgrind format.
profvis: Interactive exploration of the flame graph and call tree in a web browser.
prof.tree: Shows a call tree on the console. A similar project, treeprof, never made it to CRAN.
I’d also like to mention two other tools outside the R ecosystem:
pprof: A new tool written in Go, supports varios output formats (including callgrind) and also interactive exploration of the flame graph and call graph in a web browser.KCacheGrind: KDE-based interactive exploration, expects callgrind input.
Caveats
A few major points have been left unresolved until a forthcoming release:
- Nested calls back into R code are not fully supported yet. In situations where native code evaluates R expressions which then invoke native code again, the recorded stack trace will look wrong.
If your R code calls into native code, or if you want to try pprof with R profiling results, give the jointprof package a try! If you discover problems or have comments, file an issue. I’d be especially grateful for help with other Linux distributions or OS X.